This page is dedicated to presenting the outputs generated by work package 3 (WP3) in the Joint-Integrative Project ORION, part of the One-Health European Joint Programme.
All ORION outputs (including this and other WPs) are available in the ORION Knowledge Hub (orion.onehealthejp.eu).
[========]
WP3 focused on addressing challenges and opportunities for DATA INTEROPERABILITY in One-Health Surveillance (OHS). The final deliverable has been edited into a document of general interest in this subject. You can download it here. In this document you will find:
- a short Introduction about Linked data, data interoperability and data fairness.
- a description of the data interoperability tools developed in the project
- a description of workflows to use these tools
- a discussion about the opportunities to connect data in OHS.
The document "DATA INTEROPERABILITY in One-Health Surveillance (OHS)" presents the view of One-Health surveillance as a knowledge model, that respects data governance aspects, while connecting data in a future-proof ecosystem.
[========]
Figure 1 below depicts our understanding of the surveillance process within three specific health sectors: animal health, food safety and public (human) health, highlighting the data interoperability dimensions that emerge when collaboration and communication across these three domains is needed in order to promote One-Health.
Figure 2 then shows the WP3 activities and outputs from each of these activities.
Here you can find links to each of those resources. Visit the links to get the resources and any further documentation and details about them.
1) Health Surveillance Ontology:
1.1 dedicated page with all resources : http://datadrivensurveillance.org/health-surveillance-ontology-hso/
1.2 direct link to codes on GitHub: https://github.com/SVA-SE/HSO
1.3 direct link to codes on BioPortal: https://bioportal.bioontology.org/ontologies/HSO
1.4 permalink for access by software agents: https://www.w3id.org/hso
1.5 HSO is a member of the Open Biomedical Ontologies Foundry: http://www.obofoundry.org/ontology/hso.html
2) Excel plug-in to annotate data
2.1 Install the plug-in: https://github.com/RealEstateCore/ExcelRDF
3) Publication of the Swedish Surveillance Report as Linked-Open-Data
3.1 Specific information about Campylobacter surveillance in Sweden, which was the user case for the application of linked data principles in the examples given below: http://datadrivensurveillance.org/campylobacter-surveillance-in-sweden/
3.2 Surveillance report 2019 published as a findable and reusable resource: https://www.dataportal.se/en/datasets/59_1643/surveillance-of-infectious-diseases-in-animals-and-humans-in-sweden-2019
3.4 Campylobacter surveillance data published as a fully FAIR resource: https://www.dataportal.se/en/datasets/59_1684/campylobacter-surveillance-in-sweden
3.5 detailed description of the workflows implemented to generate FAIR data: access the "DATA INTEROPERABILITY in One-Health Surveillance (OHS)" document, referenced above.
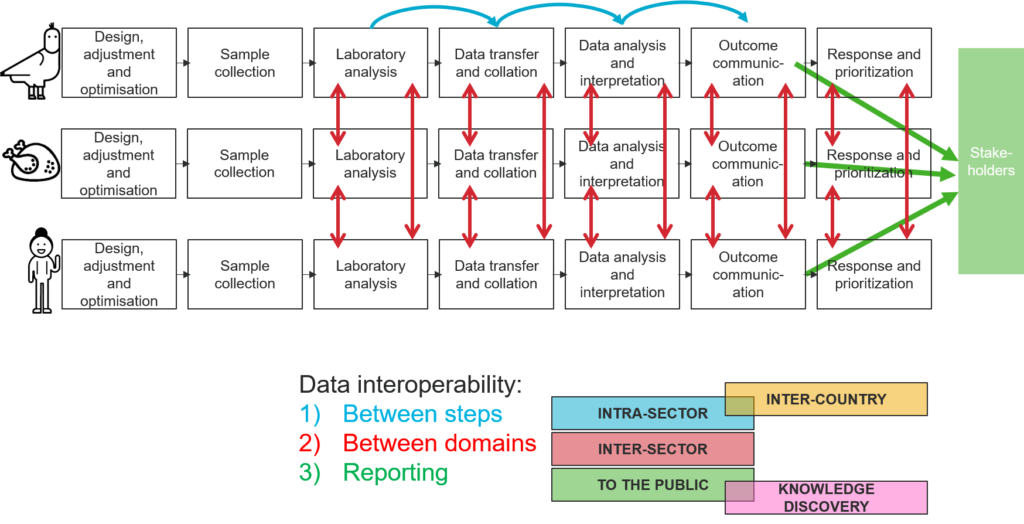
Figure 1 - Data interoperability dimensions in One-Health Surveillance.
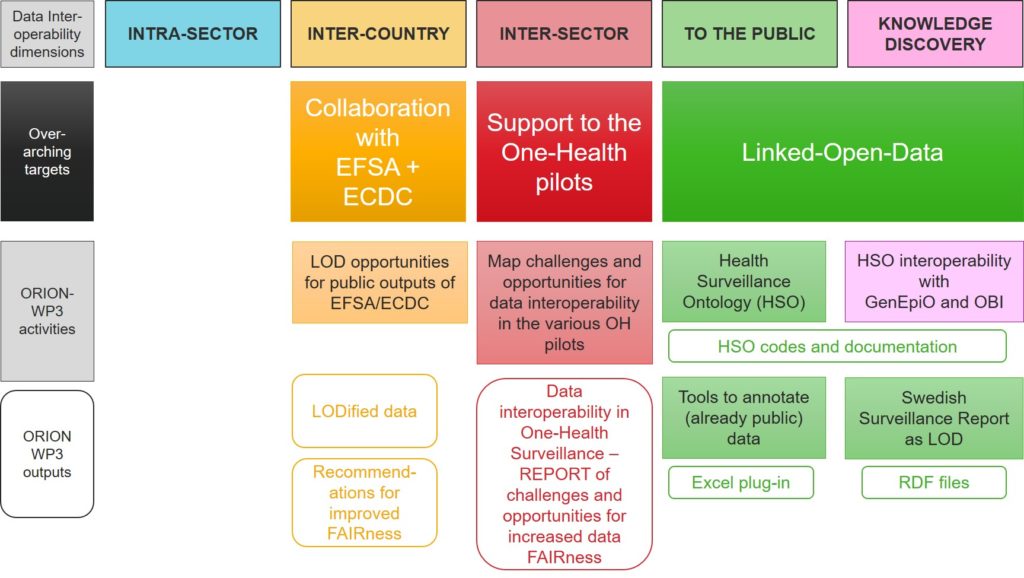
Figure 2 - ORION's WP3 activities and outputs.
Read more about the ORION project: onehealthejp.eu/orion
Read more about the OHEJP: onehealthejp.eu
The One Health European Joint Programme has received funding from the European Union’s Horizon 2020 research and innovation programme under Grant Agreement No 773830.
